Arabs account for 5% of the world population and have a high burden of cardiometabolic disease, yet clinical utility of polygenic risk prediction in Arabs remains understudied. Among 5399 Arab patients, we optimize polygenic scores for 10 cardiometabolic traits, achieving a performance that is better than published scores and on par with performance in European-ancestry individuals. Odds ratio per standard deviation (OR per SD) for a type 2 diabetes score was 1.83 (95% CI 1.74–1.92), and each SD of body mass index (BMI) score was associated with 1.18 kg/m2 difference in BMI. Polygenic scores associated with disease independent of conventional risk factors, and also associated with disease severity—OR per SD for coronary artery disease (CAD) was 1.78 (95% CI 1.66–1.90) for three-vessel CAD and 1.41 (95% CI 1.29–1.53) for one-vessel CAD. We propose a pragmatic framework leveraging public data as one way to advance equitable clinical implementation of polygenic scores in non-European populations.
Introduction
Polygenic scores can identify individuals at risk of disease, but their use in clinical practice is limited by the lack of widely accepted standards and reduced cross-ethnic transferability1,2,3,4,5. Despite many statistical methods and published scores, there is no clear framework to guide a new population interested in implementing polygenic scores using this publicly available data. Cross-ethnic transferability of scores—mostly derived from individuals of European ancestry—to other populations who are less represented in genome-wide association studies (GWAS) also suffers from reduction in performance, but new computational methods are improving on this limitation for Asian, African, and other ancestries1,2,3,4,6,7.
While the case for clinical utility of polygenic scores—mostly for cardiometabolic disease and some cancers—has been made in European-ancestry populations in the U.S. and Europe8,9,10,11, it is equally important to understand whether the prospect of clinical utility is also relevant to other populations, where genetic ancestry, environmental factors, and disease epidemiology might differ12. A recent statement from the American Society of Human Genetics highlighted the problem of reduced portability as a key priority area in human genetics research12.
Arabs represent about 5% of the world population and are massively under-represented in genomic studies worldwide, yet minimal efforts have been made to date to understand clinical utility of polygenic scores in this group13,14. Efforts to understand performance and potential utility of polygenic scores in Arabs are important for two reasons. First, Arabs represent a large and diverse ethnic group inhabiting the Middle East and North Africa, and are present as a diaspora in the United States and Western Europe13. Arab countries are also among those with the largest population growth worldwide15. Second, there is a large burden of cardiometabolic disease among Arabs with some of the countries in the Arabian peninsula having the highest rates of diabetes and obesity worldwide16,17,18,19,20,21,22,23,24. While conventional risk factors such as poor diet, smoking and sedentary lifestyle are highly prevalent, they incompletely capture excess risk; therefore, it is not clear whether genomic risk may contribute and augment risk identification.
Prior genomic studies for cardiometabolic disease in Arabs are limited due to small (i.e., typically <10,000 participants) genome-wide association studies, no prior efforts to optimize polygenic scores, and most studies being conducted on heterogeneous ethnic Arabs of diverse genetic ancestries25,26,27,28,29,30. The most comprehensive effort from the Qatar Biobank recently reported differences in linkage disequilibrium and effect sizes in a GWAS of 45 traits and showed that European-derived polygenic scores have reduced performance28. The study population represented the wider Middle Eastern region with only 37.6% “general” Arabs and 17.3% “Peninsular” Arab, but there was no analysis of score performance in those subpopulations28. In this study, we leveraged recent GWAS data and novel multi-ethnic computational methods to optimize polygenic scores for cardiometabolic disease and define their clinical utility in a cohort of indigenous Arabs from Saudi Arabia consisting of 5399 patients and 1017 population reference participants. In using publicly available data, we define a framework for optimizing polygenic scores that could be transposable to other populations.
Results
Pragmatic framework for optimizing polygenic scores to a new population
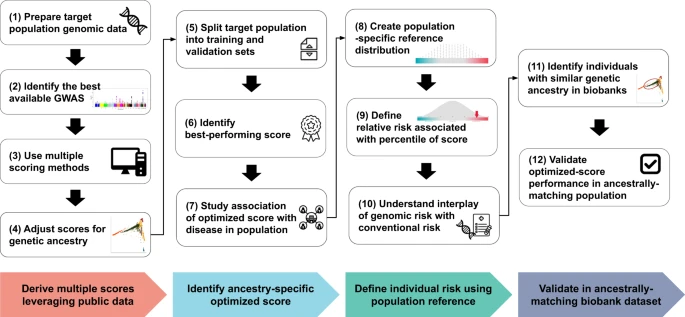
We propose a pragmatic approach to optimize polygenic scores for a new population that leverages available public datasets and recent advances in computational methods (Fig. 1). With the growth of large genomic datasets, there is an important focus on their disproportionate enrichment for individuals of European ancestry and an urgent need for more non-European representation1,14. However, less attention is paid to smaller subpopulations beyond the continental ancestries and populations with different environmental factors, where portability of polygenic scores might also be limited. Even in the absence of large genomic datasets from those target subpopulations, we show that a pragmatic framework consisting of four key steps could enable successful optimization for indigenous Arabs.
Fig. 1: Framework for optimizing polygenic scores for a new population.
A pragmatic framework for optimizing polygenic scores for a target population using publicly accessible datasets and methods consists of four steps. First, genomic data is prepared using standard quality control and imputation to obtain multiple polygenic scores based on available large and diverse GWAS results and various score derivation methods. The raw scores are adjusted for population structure using principal components (PCs) of ancestry. Second, a best-performing score is identified by splitting the dataset into training and validation sets to determine the best model in the training set and to assess the association between ancestry-specific optimized scores and traits in the validation set. Third, individual risk percentile rank is derived from the distribution based on the reference population of the same ancestry in order to identify individual relative risk levels and study the interplay between genetic risk and conventional risk factors. Fourth, to validate the ancestry-specific optimized scores, ancestry-matched samples can be identified in a large biobank dataset using genetic distance.
First, multiple scores are derived using public datasets. The target population genomic data undergoes standard quality control, and imputation for genotyping array data is performed using publicly available imputation panels, prioritizing ancestry-specific (if present) or large and diverse panels. Summary statistics from the largest and most diverse GWAS for the trait of interest are obtained, and multiple scores are derived using five scoring methods—a baseline method (PRSice-2), as well as methods that factor in the genetic architecture (LDpred2, lassosum2, and PRS-CS) and ancestry (PRS-CSx)6,31,32,33,34. A fixed set of single nucleotide polymorphisms (SNPs) from the target population is used to calculate principal components (PCs) of ancestry which are used to adjust the raw polygenic scores as described previously7,35.
Second, the target population is split into training and validation sets. The performance of the different scores for a trait is compared in the training set, and the best-performing score is selected and reported in the validation set where its association with the trait of interest is reported using regression models.
Third, since a polygenic score is best represented as percentile, we propose having a static population reference distribution on which polygenic score percentiles are defined. In the case of Arabs, we use a cohort of 1017 unrelated individuals who self-identify with 28 major clans/tribes in Saudi Arabia and are representative of the general population (Supplementary Fig. 1)36. All optimized scores are calculated in this population and used to define percentiles for each score. Subsequently, percentiles of risk are defined based on the population reference distribution, and a relative risk is assigned to each percentile of the score. Downstream analysis enables understanding of the interplay between the risk associated with genomics and conventional risk factors.
Fourth, we propose a method for identifying ancestrally matched individuals to the target sub-population in large biobanks—for example, we could identify participants in the UK Biobank who are ancestrally matched to indigenous Arabs from Saudi Arabia37. We demonstrate how such a dataset could be used to extend the optimization of polygenic scores in one country to individuals of a similar ancestry who are living in other parts of the world such as the United States or Europe.
European-derived polygenic scores have reduced performance in Arabs
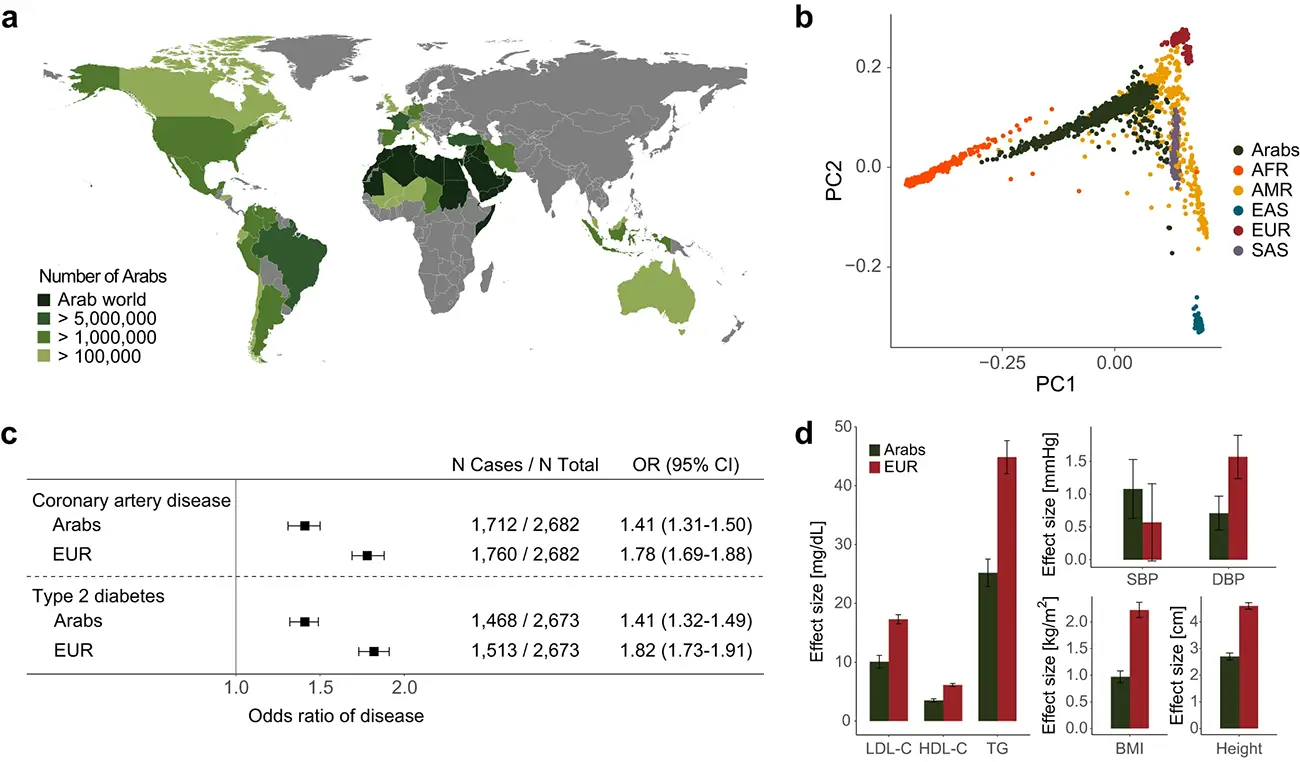
Most published polygenic scores are derived from datasets with predominantly individuals of European ancestry. In the Polygenic Score Catalog, 98.0% (1772 of 1808) published scores were trained in datasets that had at least 80% European ancestry distribution38. We first asked whether the most commonly used polygenic scores for cardiometabolic disease have reduced performance in Arabs (Fig. 2).
Fig. 2: European-derived polygenic scores have reduced performance in Arabs.
a Map showing the distribution of Arab populations around the world (created using the ‘rnaturalearth’ package in R and the cited data sources)13, 57, 58.
b Principal components of ancestry plot showing indigenous Arabs (in dark green) in this study compared to 1000 Genomes Project populations (AFR: African (in orange); AMR: Admixed American (in yellow); EAS: East Asian (in dark blue); EUR: European (in dark red); SAS: South Asian (in violet)).
c Performance of polygenic scores for coronary artery disease and type 2 diabetes in Arabs from this study vs. a matched case-control sample of European-ancestry individuals from the UK Biobank (EUR). The polygenic score for cardiomyopathy (PGS002051) has been derived in the UK Biobank where it has an inflated estimate of effect size making a comparison to Arabs inaccurate. N total is the total number of samples in the validation dataset excluding missing values for each disease. Odds ratio per standard deviation of the score is derived from a logistic regression model adjusted for age, sex, array version, and first 10 principal components of ancestry. The black boxes indicate the adjusted odds ratio. The horizontal lines around the black boxes indicate the 95% confidence intervals.
d Performance of polygenic scores for LDL cholesterol (LDL-C), HDL cholesterol (HDL-C), triglycerides (TG), systolic blood pressure (SBP), diastolic blood pressure (DBP), body mass index (BMI), and height in Arabs from this study (dark green) vs. a matched case-control sample of European-ancestry individuals from the UK Biobank (EUR, dark red). Sample sizes for all traits are included in Supplementary Table 2. Effect size estimates are derived from linear regression models adjusted for age, sex, array version, and first 10 principal components of ancestry. Error bars represent the standard error.